
How cells protect their chromosomes – the latest findings from Nature Communications
The results of research conducted by a team of scientists from University of Wrocław, recently published in Nature Communications, contribute to a deeper understanding of the processes underlying cancer cell transformation and may in the future help to better explain how DNA repair defects contribute to the development of cancer in humans.
Cells must copy and repair DNA with extreme precision to avoid damaging genetic material. Centromeres, regions responsible for the correct distribution of sister chromatids (1) between the mother cell and the daughter cell during cell division, are one of the most sensitive regions of chromosomes. Even minor abnormalities in this area can lead to serious disturbances in genome stability.
In a recently published study in Nature Communications, the team from the DNA Repair and Replication Research Center, led by dr hab. Karol Kramarz, showed that a special modification of repair proteins (known as polySUMOylation) acts as a molecular ‘brake’. It limits the rearrangements of genetic material in centromeres in fission yeast Schizosaccharomyces pombe. Thanks to this mechanism, the cell prevents the formation of undesirable changes in chromosome structure and protects genome stability. It is worth noting that this study is entirely the result of the activities of the DNA Repair and Replication Research Center with dr Katarzyna Markowska, the first author of the publication, playing a key role in the development of the project.
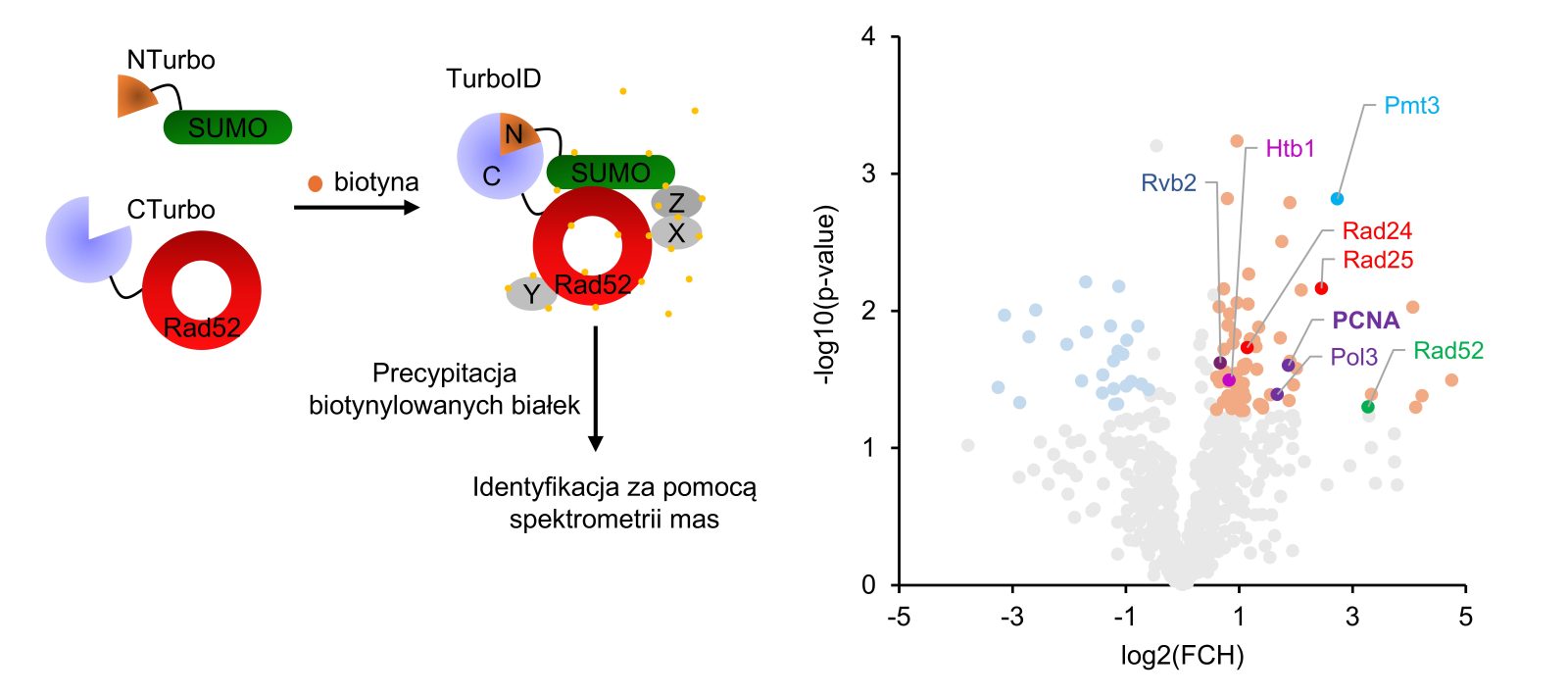
Dr hab. Karol Kramarz’s team has demonstrated new functions of the SUMO (small ubiquitin-like modifier) – a protein present in all eukaryotic organisms, including humans, which regulates cell function by (among other things) directing proteins for degradation or altering their properties and localization. SUMO is a key regulator of basic processes related to DNA metabolism, both in single-celled yeast and in human cells. Like ubiquitin, SUMO can form polymeric chains, although previously this modification was primarily associated with targeting proteins for degradation. The data obtained by the team suggested a role for SUMO chains in the organisation of the centromere structure – a region composed of numerous repeated chromosome sequences, which is a difficult to replicate site of the genome. Dr hab. Ireneusz Litwin introduced a technique for analysing the DNA structure of centromeres using next-generation sequencing into routine use, while dr Paulina Tomaszewska developed a methodology for bioinformatic analysis of the obtained data. Thanks to these tools it was possible to directly show that the lack of polySUMOylation leads to changes in centromere organisation. The study also utilised a modern proteomic technique to examine local SUMOylation, which was made possible thanks to the work of Julia Kończak, then a master’s student in the team. The composition of SUMOylated proteins involved in DNA repair was analysed by dr Michał Tracz from the Faculty of Biotechnology, who used a mass spectrometer obtained as part of the IDUB programme. This led to the identification of the PCNA (proliferating cell nuclear antigen) protein, a key replication factor. The analysis of PCNA SUMOylation was carried by doctoral student Dorota Misiorna, who also pointed out that polySUMOylation of this protein influences the activation of appropriate repair pathways that the cell uses to restart the halted DNA synthesis process. The absence of SUMO chains on PCNA leads to increased recombination within centromeres, which may disrupt their structure. Direct measurements of recombination levels in centromeres were performed by doctoral student Aleksandra Bogdańska. Finally, research conducted by Miki Haenen, a student pursuing a master’s degree as part of the Erasmus+ exchange programme, concerning the impact of an artificial SUMO chain on one of the repair proteins contributed to a better understanding of the mechanism of recombination in the centromere.
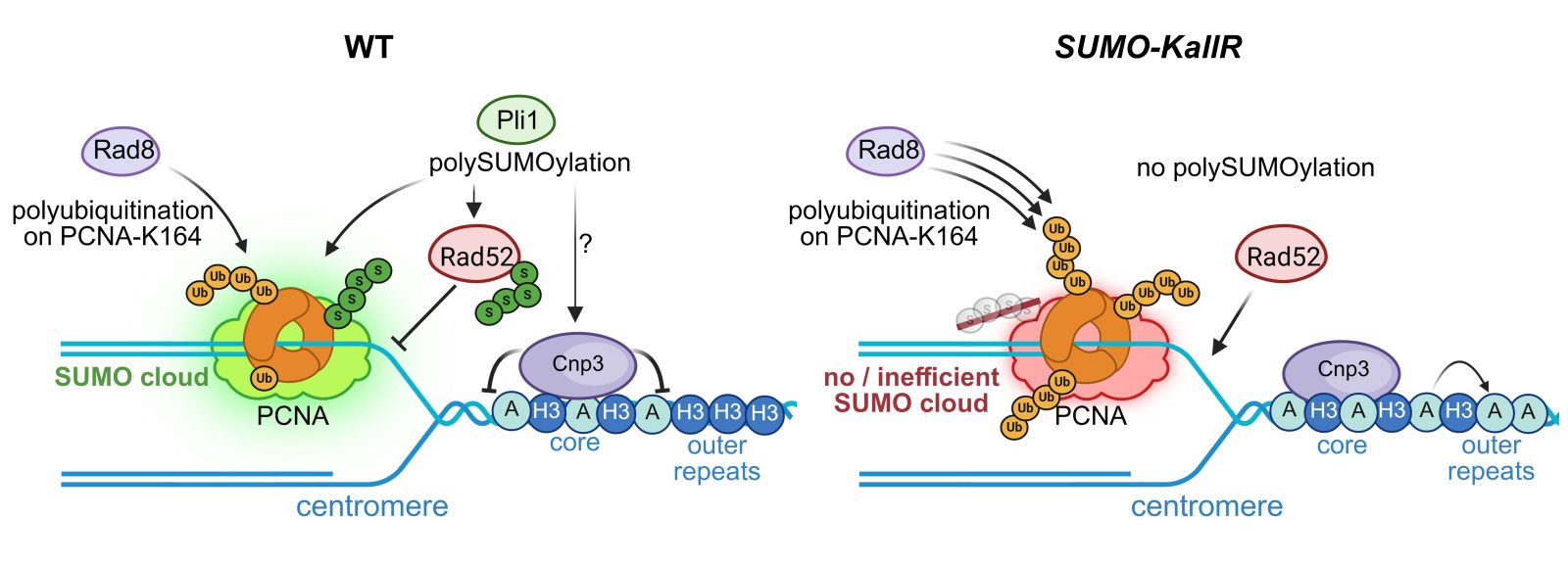
As a result, dr hab. Karol Kramarz’s team proved that polySUMOylation, previously associated mainly with directing modified proteins to the degradation pathway, plays an important role in protecting centromeres from excessive recombination. More specifically, the researchers showed that polySUMOylation of PCNA, a key protein involved in DNA replication, acts as a ‘fuse’ that prevents excessive ubiquitination of this protein. This determines how the cell resumes halted DNA synthesis in centromere region. If polySUMOylation is absent, DNA repair pathways are improperly activated, which increases recombination in centromeres and can disrupt their proper structure.
SUMO chains help maintain genome stability under normal conditions, in which cells are exposed to a constant, low level of stress, i.e. in the situation that human cells find themselves in every day. Uncontrolled recombination under normal conditions leads to genome rearrangements, which may be at the heart of cancerous transformation of human cells. Human chromosome centromeres are sensitive regions of the genome prone to frequent rearrangements. Therefore, these findings contribute to a deeper understanding of the processes underlying cancer cell transformation and may in the future help to better explain how DNA repair defects contribute to the development of cancer in humans.
Link to the publication: PolySUMOylation of PCNA and Rad52 restricts centromeric recombination in fission yeast.
***
1sister chromatids – identical copies of the same DNA molecule, formed as a result of chromosome replication, connected to each other in the centromere region.
Translated by Marta Burkiet (student of English Studies at the University of Wrocław) as part of the translation practice.
Date of publication: 10.12.2025
Added by: E.K.



